2 Data Science
Numpy
- Slice
- np.arrange(start,stop,step)
- np.linspace(start,stop,num)
- Random
- uniform: np.random.randint(low,high,size)
- standard normal: np.random.randn(d1,d2,...)
- max,min
- argmax, argmin return the indices of the maximum/minimum values
Ndarray
numpy shape is determined by the iteration of square brackets
a = np.shape(1) # shape:()
a = np.array([1, 2, 3, 4]) # shape: (4,)
a = a[:,np.newaxis] # array([[0],[1],[2]]),shape: (4,1)
b = np.shape([[1, 3]]) # (1,2)
c = np.array([(1,2),(3,4),(5,5),(423,12)],dtype=[('x', 'i4'), ('y', 'i4')])
print(c.shape, c.ndim) # (4,) 1
d = np.array([(1,2),(3,4),(5,5),(423,12)]) # shape: (4,2) ndim:2
indexing
## Elipsis
a = np.arange(120).reshape(2, 3, 4, 5)
print(a[:, :, :, 0])
print(a[..., 0]) # same
# [[[ 0 5 10 15]
# [ 20 25 30 35]
# [ 40 45 50 55]]
#
# [[ 60 65 70 75]
# [ 80 85 90 95]
# [100 105 110 115]]]
print(a[0, :, :, 0])
print(a[0, ..., 0]) # same
Basic operation
elementwise vs dot
a = np.array([20, 30, 40, 50])
b = np.arange(4)
c = a + b # Matrix
print(c) # [20 31 42 53]
print(a*b) # same as np.multiply(): [0 30 80 150]
print(a.dot(b)) # same as np.matmul(): 260
With the larger dataset, we see that using NumPy results in code that executes over 50 times faster! Throughout this course (and in the real world), you will find that writing efficient code will be important; arrays and vectorized operations are the most common way of making Python programs run quickly.
Advanced Indexing
Index by arrays of indices
a = np.arange(12)**2 # the first 12 square numbers
i = np.array([1, 1, 3, 8, 5]) # an array of indices
print(a[i]) # array([ 1, 1, 9, 64, 25])
j = np.array([[3, 4], [9, 7]]) # a bidimensional array of indices
print(a[j]) # array([[ 9, 16],[81, 49]])
Index by boolean arrays
a = np.arange(12).reshape(3, 4)
a[a>4] = 0 # 0 is a matrix
print(a) # array([[0, 1, 2, 3],[4, 0, 0, 0],[0, 0, 0, 0]])
Deep copy
If a = b, that's only a reference
a = np.arange(int(1e8))
b = a[:100].copy()
del a # the memory of ``a`` can be released.
Pandas
- pd.Series(data,index): One-dimensional ndarray with axis labels. data could be array-like, Iterable, dict, or scalar value
- pd.DataFrame(data,index,column): Two-dimensional, size-mutable, potentially heterogeneous tabular data.
Selection
- Select by column
- Slice the row
- Select by label
df = pd.DataFrame(
{
"A": 1.0,
"B": pd.Timestamp("20130102"),
"C": pd.Series(1, index=list(range(4)), dtype="float32"),
"D": np.array([3] * 4, dtype="int32"),
"E": pd.Categorical(["test", "train", "test", "train"]),
"F": "foo",
}
)
## 1. Select by column
print(df['A'])
# 0 1.0
# 1 1.0
# 2 1.0
# 3 1.0
## 2. Slice the row
print(df[0:2])
# A B C D E F
# 0 1.0 2013-01-02 1.0 3 test foo
# 1 1.0 2013-01-02 1.0 3 train foo
## 3. Select by label
print(df.loc[[0,2],['A','B']])
# Name: A, dtype: float64
# A B
# 0 1.0 2013-01-02
# 2 1.0 2013-01-02
MultiIndex
tuples = list(
zip(
["bar", "bar", "baz", "baz", "foo", "foo", "qux", "qux"],
["one", "two", "one", "two", "one", "two", "one", "two"],
)
)
index = pd.MultiIndex.from_tuples(tuples, names=["first", "second"])
df = pd.DataFrame(np.random.randn(8, 2), index=index, columns=["A", "B"])
# A B
# first second
# bar one 1.036623 0.775701
# two -0.491414 -0.596697
# baz one -1.039759 0.274683
# two -0.138051 -1.128008
# foo one -0.525837 -1.043091
# two 0.152277 1.281915
# qux one 0.625832 1.662615
# two -0.933067 -0.257373
print(df.loc['bar'].loc['one','B']) # 0.775701188641088
print(df.groupby('second').describe()['A'])
# count mean std min 25% 50% 75% max
# second
# one 4.0 0.523990 0.743125 -0.007540 0.125254 0.241141 0.639877 1.621220
# two 4.0 0.264113 0.644908 -0.414561 -0.175321 0.222076 0.661510 1.026859
Boolean Indexing
Selecting values from a DataFrame where a boolean condition is met
df = pd.DataFrame(np.random.randn(6, 4), index=pd.date_range("20130101", periods=6), columns=list("ABCD"))
print(df[df["A"] > 0])
# A B C D
# 2013-01-02 0.945462 -0.880120 0.030657 -1.906303
# 2013-01-06 0.290602 -0.155123 0.174085 -1.606480
Operations
SQL join
left = pd.DataFrame({"key": ["foo", "foo"], "lval": [1, 2]})
right = pd.DataFrame({"key": ["foo", "foo"], "rval": [4, 5]})
print(pd.merge(left,right,on = 'key'))
# key lval rval
# 0 foo 1 4
# 1 foo 1 5
# 2 foo 2 4
# 3 foo 2 5
Matplotlib
The Figure keeps track of all the child Axes, an area where points can be specified in terms of x-y coordinates
fig, axes = plt.subplots()
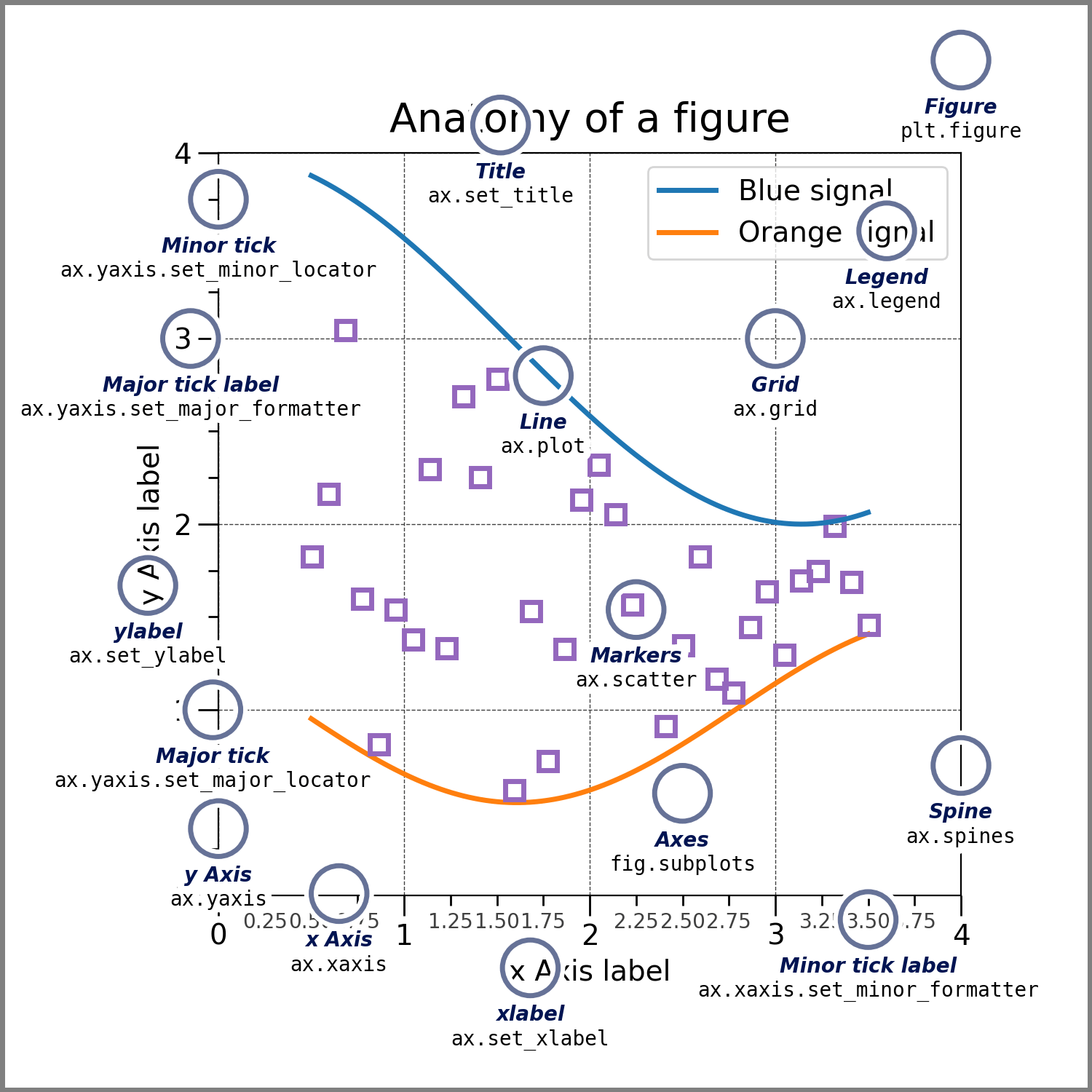
plt.tight_layout()
fig, axes = plt.subplots(1,2,figsize=(8,2))
axes[0].plot(x,y1,'b:',x,y2,'g-')
axes[0].legend(['fig1','fig2'],loc='lower left')
axes[0].set_xlabel('x label')
plt.show()
Seaborn
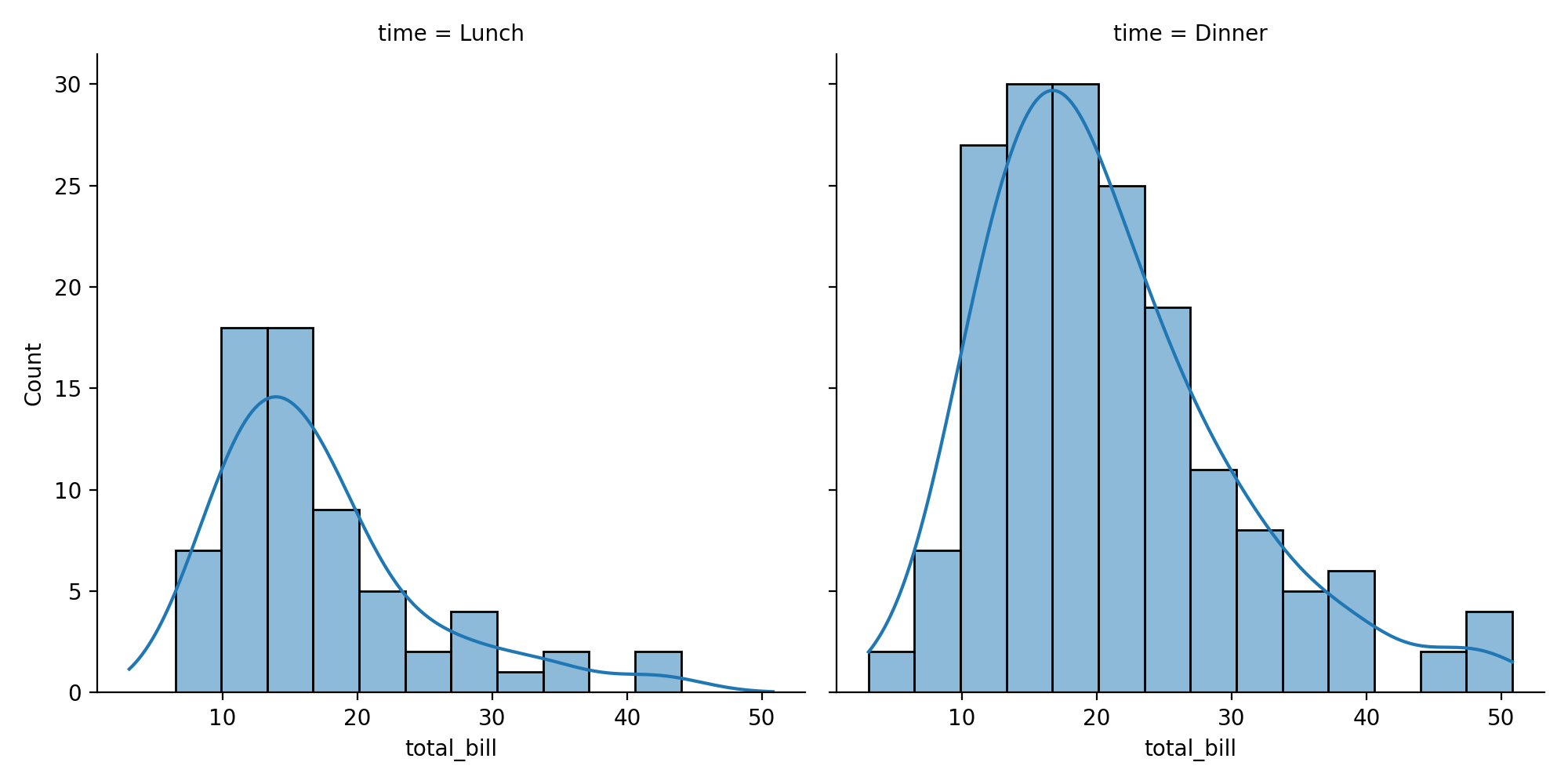
Distributional representations
KDE: kernel distribution estimation
tips = sns.load_dataset("tips")
sns.displot(data=tips, x="total_bill", col="time", kde=True)
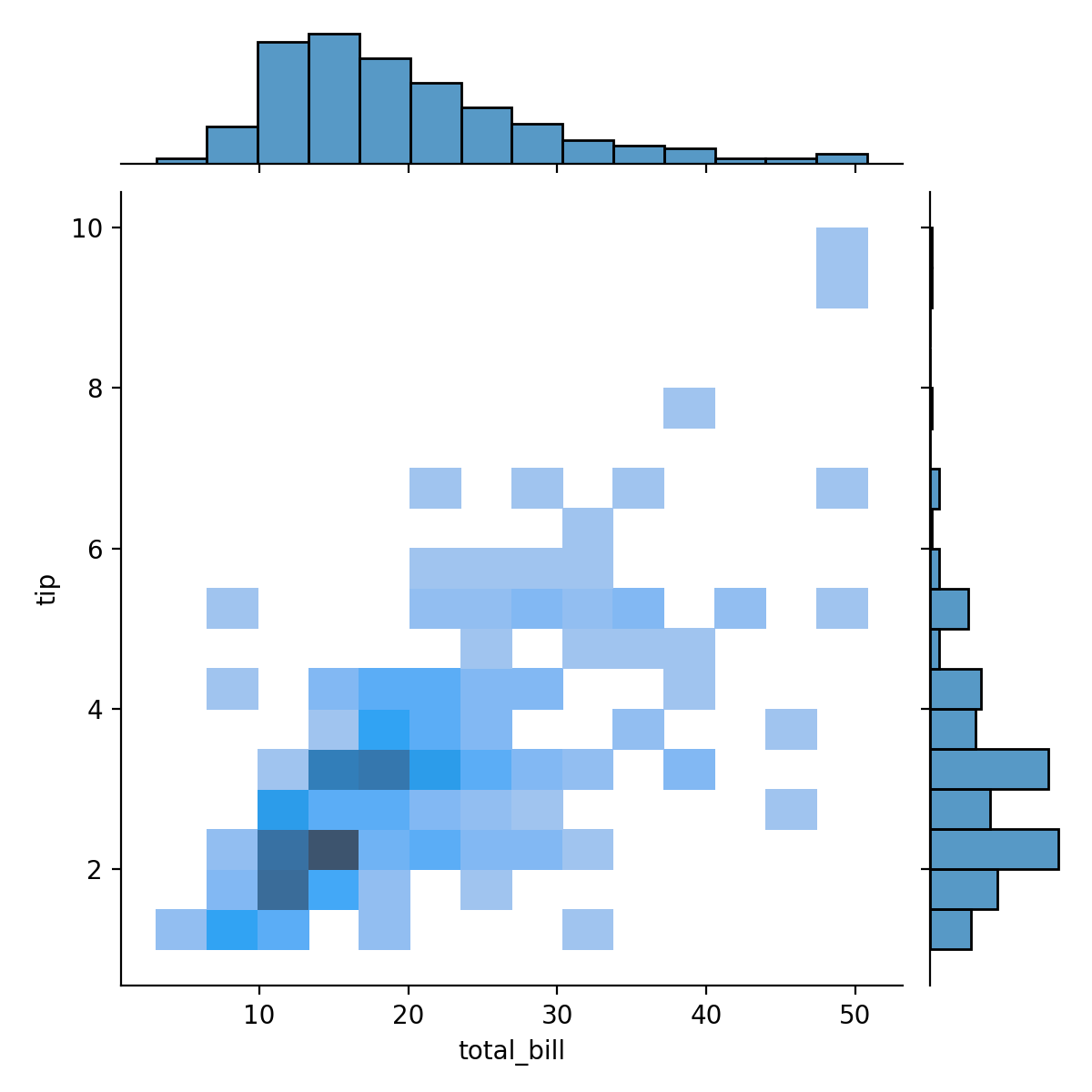
sns.jointplot(x='total_bill',y='tip',data=tips,kind="hist")
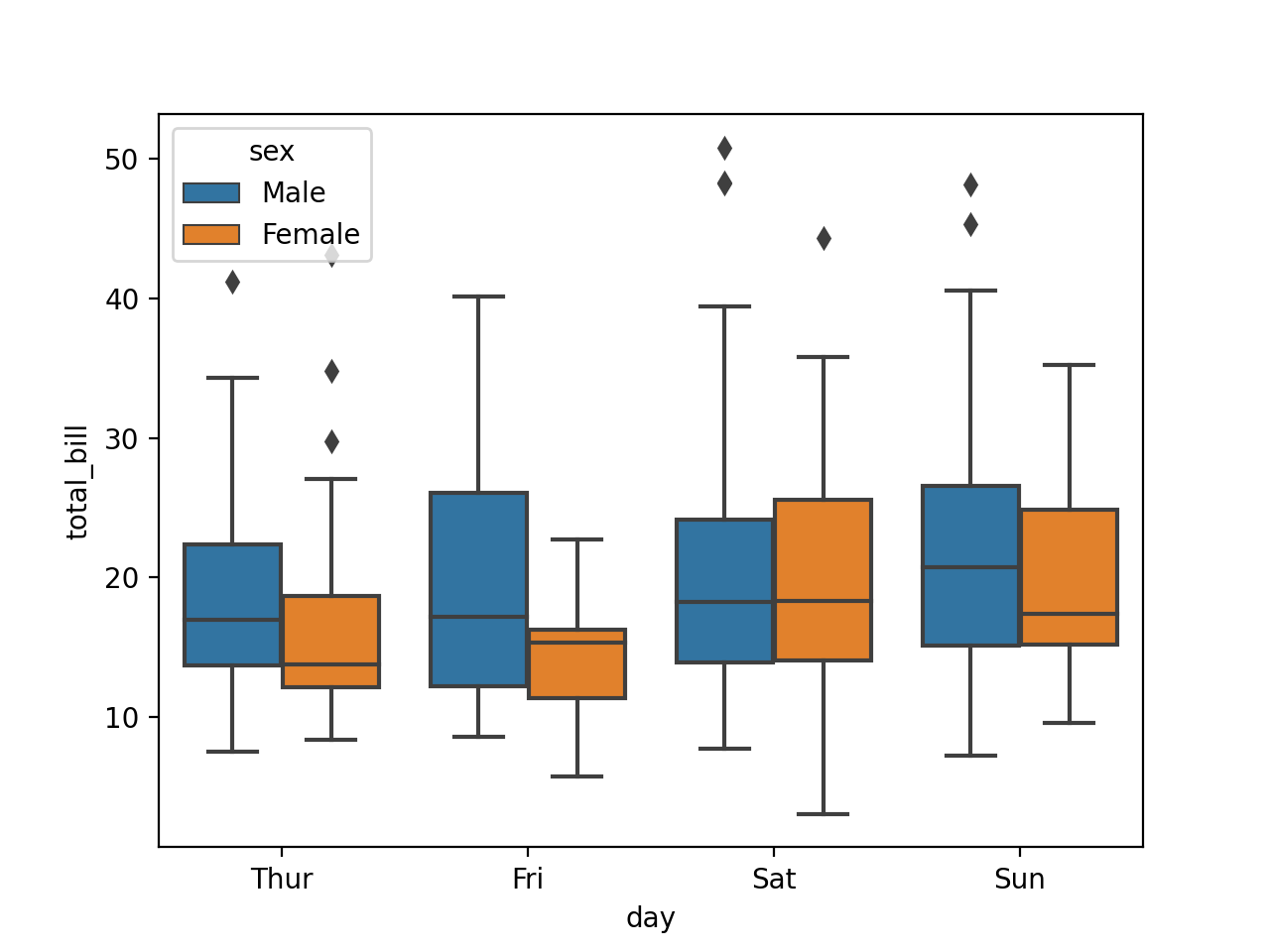
Catergorical Plot
tips = sns.load_dataset("tips")
sns.boxplot(x='day',y='total_bill',data=tips,hue='sex')

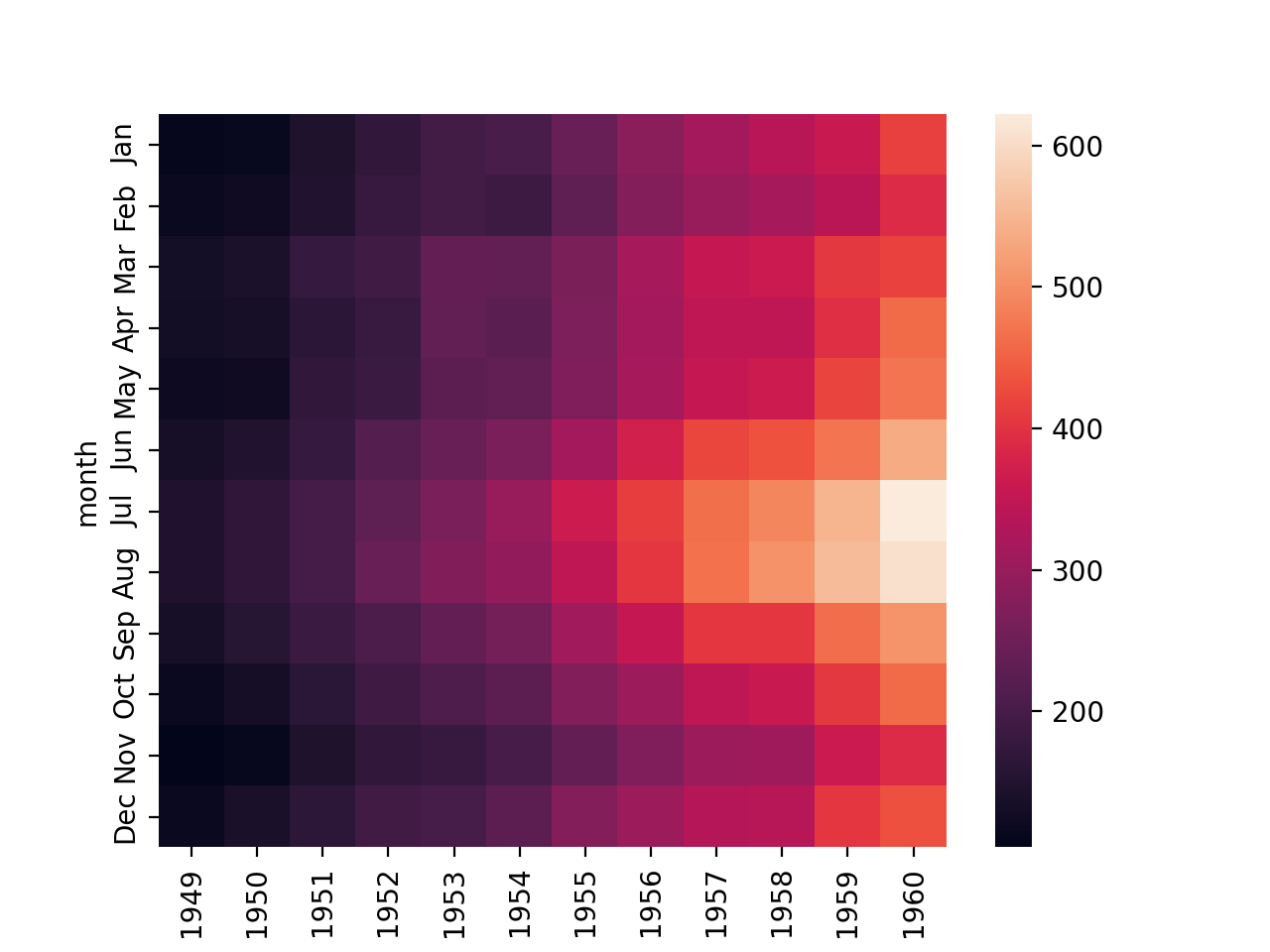
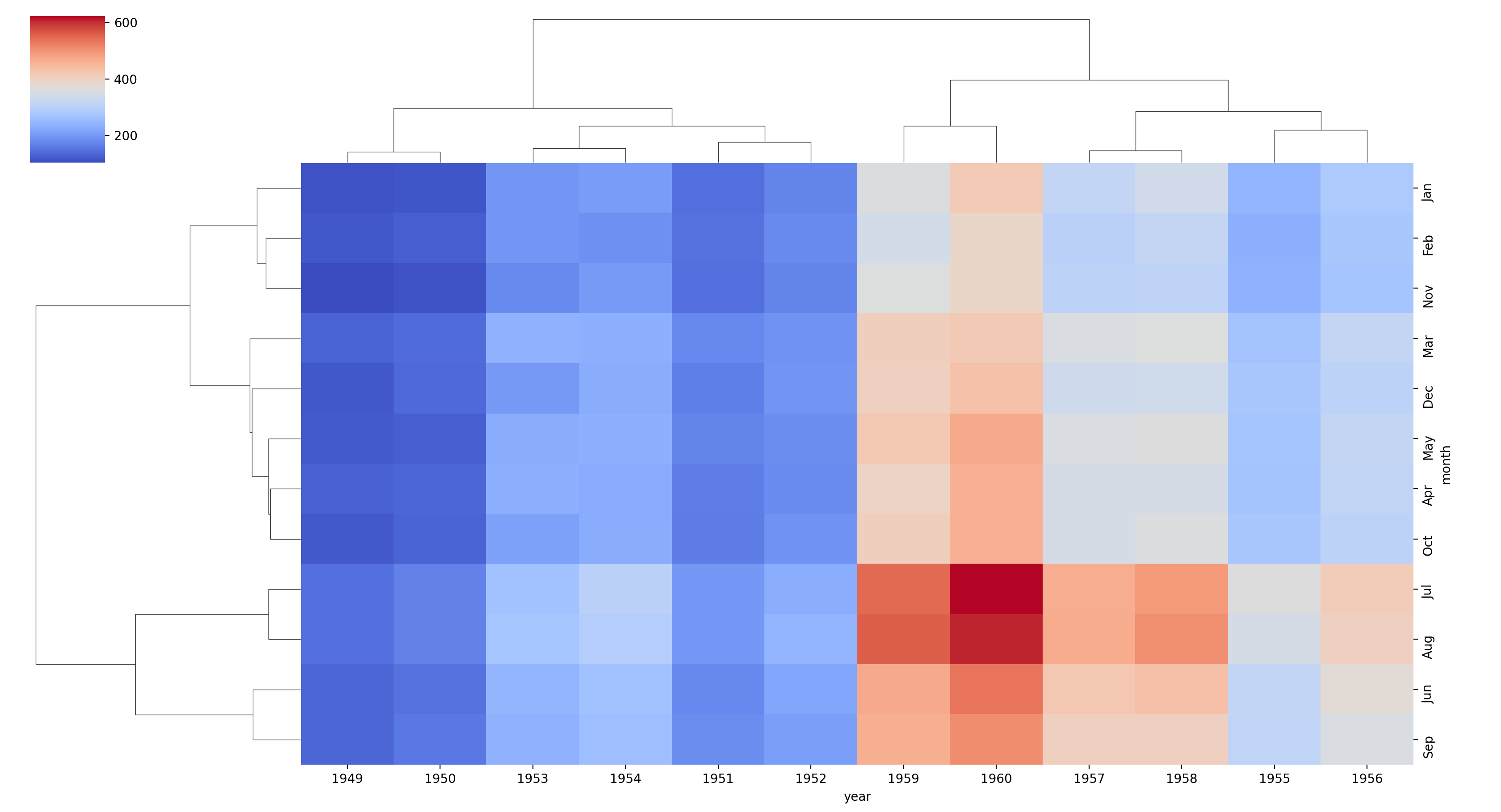
Matrix Plot
fp = flights.pivot_table(index='month',columns='year',values='passengers')
sns.heatmap(fp)
sns.clustermap(fp,cmap='coolwarm')